Nicholas Clark

PhD candidate
University of Cincinnati
I am a PhD Candidate in Biostatistics and Bioinformatics at the University of Cincinnati. I'm working on mathematical methods for predicting biological pathway perturbations in genomics data. I have also developed a few web applications in collaboration with HiTS, the Harvard Program in Therapeutic Science at Harvard Medical School.
Projects
Projects
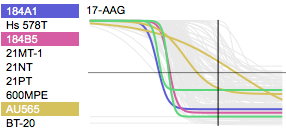
HMS LINCS Small Molecule Library Apps
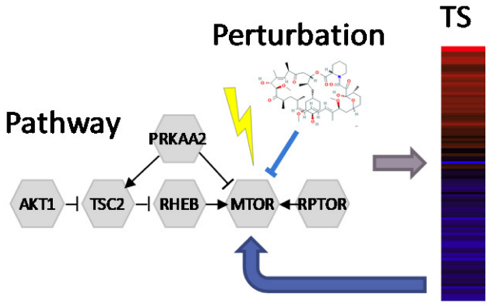
Pathway activity signatures
Software development
Selected Publications
First author
Clark, N. A. et al. GRcalculator: an online tool for calculating and mining dose–response data. BMC Cancer 17, 698 (2017). doi: 10.1186/s12885-017-3689-3.
Supporting author
Stefely, J. A. et al. Mass spectrometry proteomics reveals a function for mammalian CALCOCO1 in MTOR-regulated selective autophagy. Autophagy 1–19 (2020) doi:10.1080/15548627.2020.1719746.
Moret, N. et al. Cheminformatics Tools for Analyzing and Designing Optimized Small-Molecule Collections and Libraries. Cell Chem Biol (2019) doi:10.1016/j.chembiol.2019.02.018.
Ren, Y. et al. Predicting mechanism of action of cellular perturbations with pathway activity signatures. Biorxiv (2019) doi:10.1101/705228.
View all publications on ORCiD.